24 R Merging Exercise
24.1 Math Art made with code

24.2 Merging Best Practice
- Always be careful when merging.
- Always check for duplicated IDs before doing the merge.
- Always check that your ID columns do not contain any missing values.
- Check that the values in the ID columns (e.g., the keys) match.
- Can use an
anti_jointo check this.
- Can use an
- Inconsistencies in the values of the keys can be hard to fix.
- Always check the dimensions, before and after the merge, to make sure the merged object has the expected number of rows and columns.
- Always explicitly name the keys you are merging on.
- When using tidyverse
joincommands, load thetidylogR package in order to turn on very useful additional feedback.
24.3 Load Libraries
24.4 Input data
Let’s load the synthetic simulated Project 1 data and associated data dictionary:
24.5 Select a subset of subject-level fields
Set up a data frame a that has these subject-level fields: “subject_id” “maternal_age_delivery” “case_control_status” “prepregnancy_BMI”
24.6 Unique records
The data were given to us in a way that repeated subject-level information, once for each sample from each individual subject.
From your data frame a select only the unique records, creating data frame b.
Use the unique command from base R or the distinct command from the dplyr R package.
24.6.1 Comment
It is better to apply unique to the whole data frame, not just to the subject_id column, as that ensures that you are selecting whole records that are unique across all of their columns.
Note that the dplyr R package provides the distinct command, which keeps only unique/distinct rows from a data frame. It is faster than the unique command.
24.7 Check that the subject_id’s are now not duplicated
Are the subject_id’s unique?
Hint: In the R Recoding Reshaping Exercise you learned how to check for duplicated values in a vector. Use that knowledge here.
24.8 Create random integer IDs
Create a new column ID containing randomly chosen integer IDs; this is necessary to de-identify the data. To do this, use the sample command, sampling integers from 1 to the number of rows in data frame b.
This could also be done using the sample-int() function:
24.9 Merging Best Practice
Always be careful when merging.
- Always check for duplicated IDs before doing the merge.
- Always check that your ID columns do not contain any missing values.
- Check that the values in the ID columns (e.g., the keys) match.
- Can use an ‘anti_join’ to check this.
- Inconsistencies in the values of the keys can be hard to fix.
- Always check the dimensions to make sure the merged object has the expected number of rows and columns.
- Always explicitly name the keys you are merging on.
- If you don’t name them, then the join command will use all variables in common across
xandy.
- If you don’t name them, then the join command will use all variables in common across
- When using tidyverse
joincommands, load thetidylogR package in order to turn on very useful additional feedback.
24.10 Tidyverse join commands
Here is a nice illustration of joins from RStudio (which they shared under a CC_BY 4.0 license):
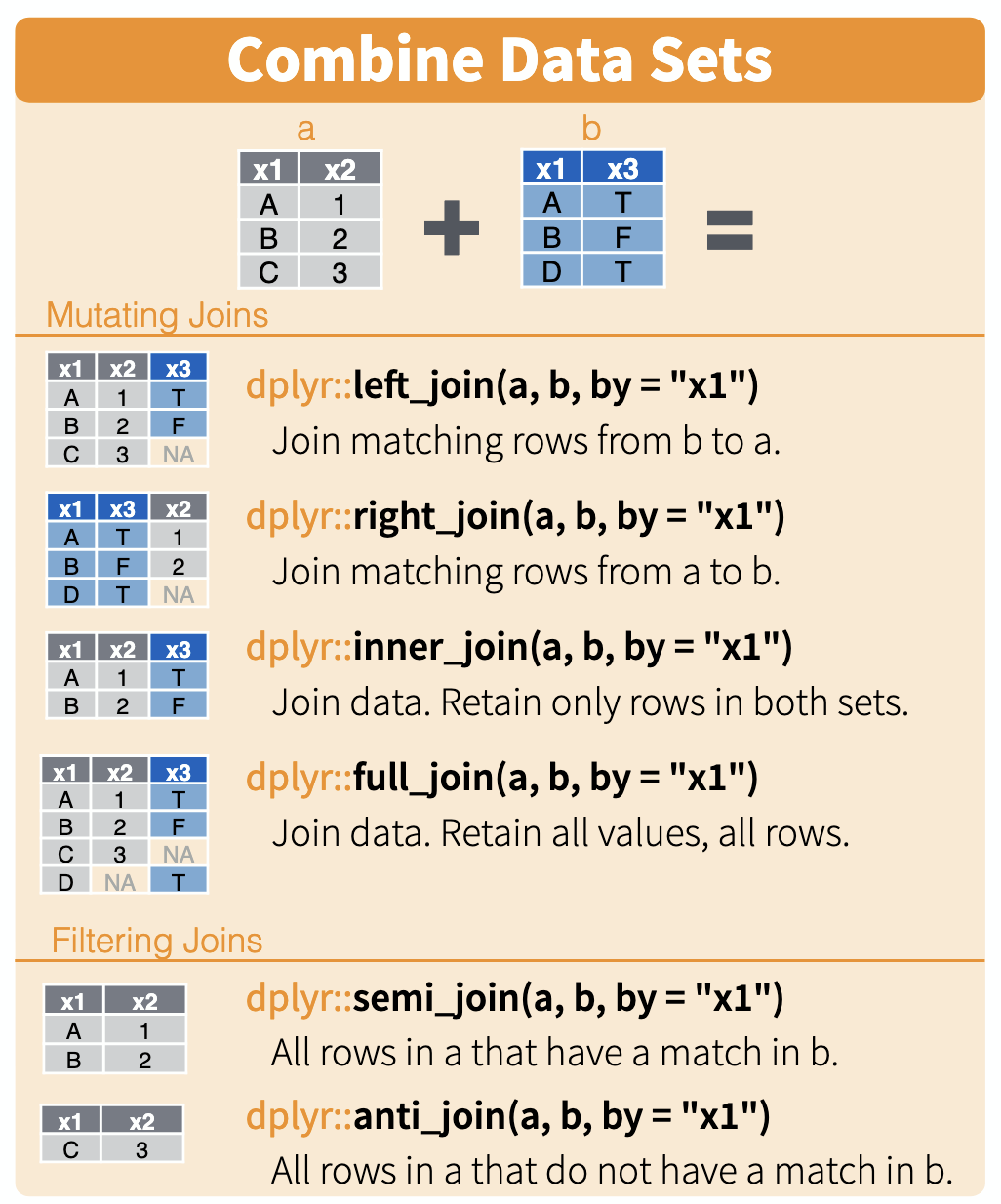
To illustrate the join commands, let’s set up these data frames a and b:
The left_join(a,b) command from the ‘tidyverse’ package keeps all observations in a, joining in matching rows from b.
The right_join(a,b) command keeps all observations in b, joining in matching rows from a.
The full_join(a,b) command keeps all observations in a and b.
The inner_join(a,b) command only keeps observations from a that have a matching key in b.
To illustrate how to handle different names for the common ID field, let’s set up these data frames a and b slightly differently:
What if one data frame has non-unique IDs?
merge: non-unique IDs in data frame b. A one-to-many relationship.
So data frame a originally had 3 records, but now the full join outputs a 5 record data frame.
merge: non-unique IDs in both data frames a and b. All possible combinations.
A filtering semi_join(a,b) returns all rows from a that have a match in b
A filtering anti_join(a,b) returns all rows from a that do not have a match in b. Useful for understanding causes of mismatches when joining.
In ‘R for Data Science’, the authors argue that using tidyverse-style commands (e.g., left_join) is better than using the merge command of base R because
“they more clearly convey the intent of your code: the difference between the joins is really important but concealed in the arguments of
merge(). dplyr’s joins are considerably faster and don’t mess with the order of the rows.”
See http://r4ds.had.co.nz/relational-data.html#other-implementations
24.11 Merge in new phenotype information
The PI has sent you new trait data for your subjects.
Carefully merge in the new data in using tidyverse commands. As this is subject-level information, it should be merged into the subject-level data frame b which was created above when from your data frame a you selected only the unique records. Following recommended best merging practice, be sure to:
- Check for duplicated IDs before doing the merge.
- Check that your ID columns do not contain any missing values.
- Check that the values in the ID columns (e.g., the keys) match.
- Check the dimensions before and after the merge, to make sure the merged object has the expected number of rows and columns.
- Explicitly name the keys you are merging on.
If you notice any problems with this merge, prepare a report for the PI detailing what you noticed and what you’d like to ask the PI about.
Here we load the tidylog R package, which will result in useful feedback when tidyverse commands are executed.
Which subject_id’s are duplicated?
Now check if there are any missing values in the ID columns?
How many of rows would you expect the merged data frame to have?
Why did the number of rows increase when doing the full join?
24.12 Further checks
When merging data based on an ID shared in common, it is not only important to check for duplicated IDs, but it is also important to check for overlap of the two ID sets.
Check if the set of subject_id IDs in your dataframe b fully overlaps the set of subject_id IDs in the new data set. If there is not full overlap, document which IDs do not overlap.
Hint: Use an anti_join.
anti_join() return all rows from x without a match in y.
24.13 Letter to the PI
Prepare a brief letter to the PI describing the problems in the data that you discovered when trying to merge in the new trait data.
Dear Dr. PI,
When trying to merge in the new trait data that you sent, I noticed a few issues that I wanted to bring to your attention.
The new data set has a duplicated record for subject_id SUBJ09.
The new data set has a subject_id that is not observed in our original data set: SUBJ00
The new data set is missing records for these two subject_id’s that are observed in our original data set: SUBJ18 and SUBJ24.
Please resolve these issues and send us a corrected version of the new trait data.
Thank you!