30 Data Cleaning Exercise
30.1 Data cleaning principles
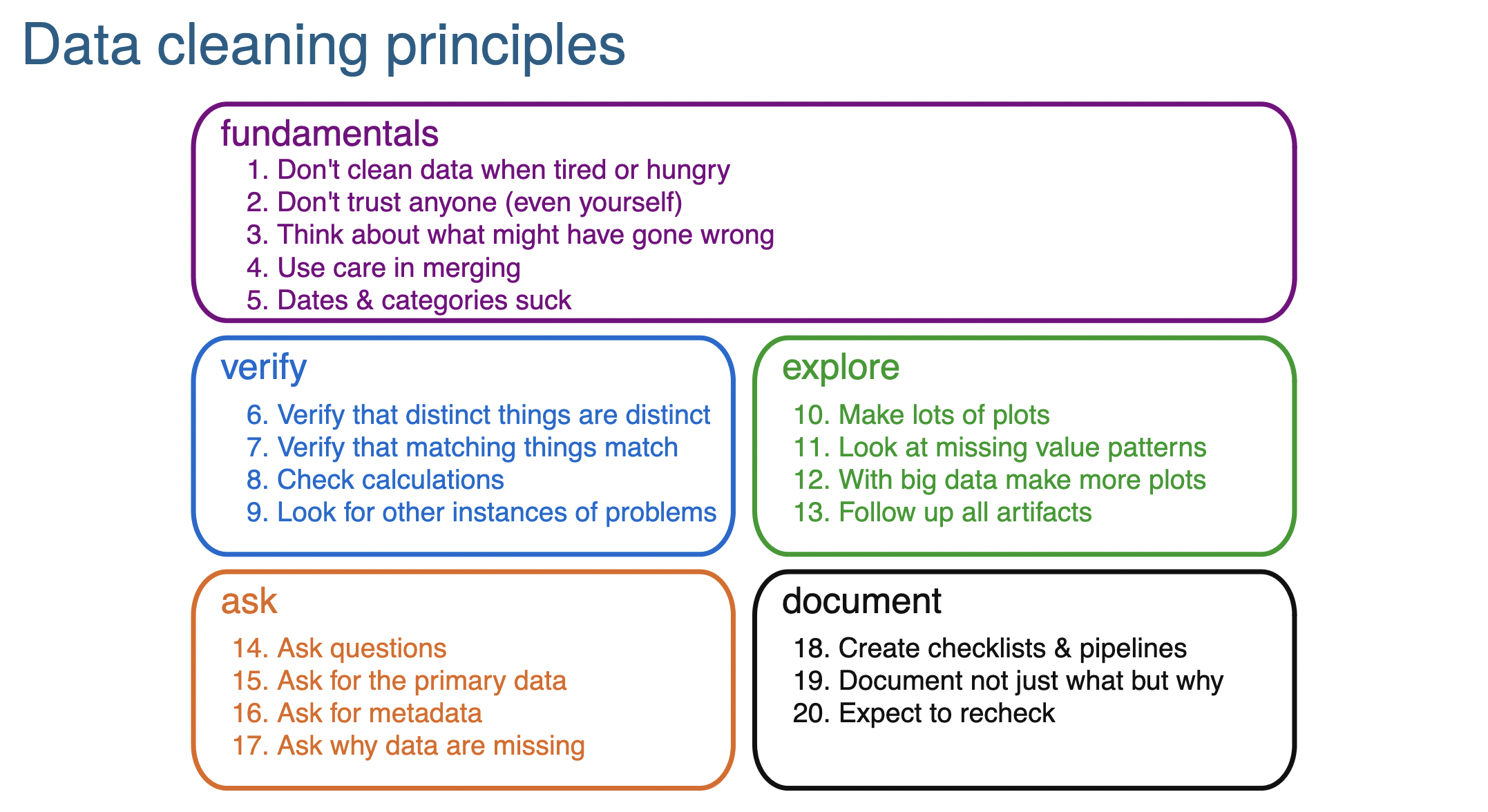
Data cleaning principles from Karl Broman’s slide set:
https://kbroman.org/Talk_DataCleaning/data_cleaning.pdf
30.2 dbGaP quality control
As described in:
Tryka KA, Hao L, Sturcke A, Jin Y, Wang ZY, Ziyabari L, Lee M, Popova N, Sharopova N, Kimura M, Feolo M. NCBI’s Database of Genotypes and Phenotypes: dbGaP. Nucleic Acids Research. 2014 Jan 1;42(D1):D975–D979. PMID: 24297256 PMCID: PMC3965052 DOI: https://doi.org/10.1093/nar/gkt1211
“The Database of Genotypes and Phenotypes (dbGap, http://www.ncbi.nlm.nih.gov/gap) is a National Institutes of Health-sponsored repository charged to archive, curate and distribute information produced by studies investigating the interaction of genotype and phenotype.”
Under NIH data sharing guidelines, all properly consented large-scale genetic or ’omics studies must deposit their data in dbGaP. To so, one must closely follow the formatting requirements as described in the dbGaP Submission Guide:
https://www.ncbi.nlm.nih.gov/gap/docs/submissionguide/
This involves setting up a data dictionary that follows dbGaP specifications:

30.3 Minimum and Maximum Values Check
30.3.1 MIN, MAX check
In the data dictionary, for some variables, MIN and MAX values may be specified. For example, for age, it has a natural minimum of zero.
MIN The logical minimum value of the variable. If a separate code such as -1 is used for a missing field, this should not be considered as the MIN value.
MAX The logical maximum value for the variable. If a separate code such as 9999 is used for a missing field, this should not be considered as the MAX value.Task: Design and implement a check that the specified MIN and MAX values observed in the data are consistent with the values as specified in the data dictionary.
Using DD.dict.I and DS.data.I, check the PERCEIVED_CONFLICT variable to see if all the values fall within the stated MIN and MAX values.
30.3.2 Pseudo-code
First try to write out an algorithm for this Minimum and Maximum Values Check in pseudo-code, outlining each step.
Possible steps:
- Read the vector of
PERCEIVED_CONFLICTfromDS.data.I - Read the
MINandMAXvalues forPERCEIVED_CONFLICTfromDD.dict.I - Count and list any
PERCEIVED_CONFLICTvalues that lie outside of the range [MIN, …,MAX].
30.3.3 Implement MIN, MAX check in R
Implement your algorithm in code.
These results are consistent with those returned by the minmax_check function from the dbGaPCheckup R package:
30.3.4 Make your check more robust
After implementing your algorithm in R code, think about it a bit further - is it robust to the situation where only one of the MIN and MAX values is specified and the other is missing? Is it robust to the situation where both MIN and MAX are missing?
The code proposed here is not robust to MIN or MAX being NA because, for example, if MIN is NA and MAX is 15, in some situations the logical indexing into the trait vector used of trait < min.val | trait > max.val would return NA instead of TRUE or FALSE as intended.
This are possible steps toward writing a more robust check:
But what would the above code return if both MIN and MAX were NA?
If we look at the minmax_check code by typing minmax_check at the R prompt, we see that it uses a which when it tries to find the out-of-range values:
flagged <- dataset_na[which(dataset_na[, ind] <
range_dictionary[1] | dataset_na[, ind] >
range_dictionary[2]), , drop = FALSE]Why is this robust to either one or both of MIN and MAX being missing?
30.3.5 Check the PREGNANT variable
Now apply your MIN and MAX checking algorithm to the PREGNANT variable.
These out-of-range values of -9999 and -4444 look kind of strange and are unexpected given the first two entries of the VALUES column of the data dictionary for this variable:
Based on this, we’d expect to see only 0 and 1 values in the PREGNANT variable.
What’s going on?
Let’s examine some more columns of the data dictionary:
30.3.6 Handle missing values
If we further check the data dictionary for the PREGNANT variable, we see that the out-of-range values we observed in our check above are actually missing value codes and so should not be flagged as being out of range.
Extend your algorithm to handle missing value codes. To do this first outline your approach in pseudo-code. Then implement it in R.
Possible steps:
- Read the vector of
PREGNANTfromDS.data.I - Read the
MINandMAXvalues forPREGNANTfromDD.dict.I - Have the user provide a list of missing value codes
- Recode any
PREGNANTvalue that matches one of the missing value codes to the standardNAR missing value code. - Count and list any non-missing
PREGNANTvalues that lie outside of the range [MIN, …,MAX].
This is essentially the approach used in the minmax_check function of the dbGaPCheckup R package.
If we examine the minmax_check code by typing minmax_check without parentheses at the R prompt, we see that this is how the missing value recoding step is done:
for (value in na.omit(non.NA.missing.codes)) {
dataset_na <- dataset_na %>% mutate(across(everything(),
~na_if(.x, value)))
}30.4 References and Resources
Heinsberg LW, Weeks DE. dbGaPCheckup: pre-submission checks of dbGaP-formatted subject phenotype files. BMC Bioinformatics. 2023 Mar 3;24(1):77. PMID: 36869285 PMCID: PMC9985192 DOI: https://doi.org/10.1186/s12859-023-05200-8
Tryka KA, Hao L, Sturcke A, Jin Y, Wang ZY, Ziyabari L, Lee M, Popova N, Sharopova N, Kimura M, Feolo M. NCBI’s Database of Genotypes and Phenotypes: dbGaP. Nucleic Acids Research. 2014 Jan 1;42(D1):D975–D979. PMID: 24297256 PMCID: PMC3965052 DOI: https://doi.org/10.1093/nar/gkt1211
dbGaPCheckup: https://lwheinsberg.github.io/dbGaPCheckup/index.html
NCBI’s GaPTools: https://github.com/ncbi/gaptools