library(tidyverse)
# library(tidylog)22 R Tidyverse Exercise
22.1 Load Libraries
Load the tidyverse packages
22.2 Untidy data
Let’s use the World Health Organization TB data set from the tidyr package
who <- tidyr::who
dim(who)[1] 7240 60head(who[,1:6] %>% filter(!is.na(new_sp_m014)))# A tibble: 6 × 6
country iso2 iso3 year new_sp_m014 new_sp_m1524
<chr> <chr> <chr> <dbl> <dbl> <dbl>
1 Afghanistan AF AFG 1997 0 10
2 Afghanistan AF AFG 1998 30 129
3 Afghanistan AF AFG 1999 8 55
4 Afghanistan AF AFG 2000 52 228
5 Afghanistan AF AFG 2001 129 379
6 Afghanistan AF AFG 2002 90 476See the help page for who for more information about this data set.
In particular, note this description:
“The data uses the original codes given by the World Health Organization. The column names for columns five through 60 are made by combining new_ to a code for method of diagnosis (rel = relapse, sn = negative pulmonary smear, sp = positive pulmonary smear, ep = extrapulmonary) to a code for gender (f = female, m = male) to a code for age group (014 = 0-14 yrs of age, 1524 = 15-24 years of age, 2534 = 25 to 34 years of age, 3544 = 35 to 44 years of age, 4554 = 45 to 54 years of age, 5564 = 55 to 64 years of age, 65 = 65 years of age or older).”
So new_sp_m014 represents the counts of new TB cases detected by a positive pulmonary smear in males in the 0-14 age group.
22.3 Tidy data
Tidy data: Have each variable in a column.
Here is a nice illustration from RStudio (which they shared under a CC_BY 4.0 license):

Question: Are these data tidy?
No these data are not tidy because aspects of the data that should be variables are encoded in the name of the variables.
These aspects are
- test type.
- sex of the subjects.
- age range of the subjects.
Question: How would we make these data tidy?
Consider this portion of the data:
head(who[,1:5] %>% filter(!is.na(new_sp_m014) & new_sp_m014>0), 1)# A tibble: 1 × 5
country iso2 iso3 year new_sp_m014
<chr> <chr> <chr> <dbl> <dbl>
1 Afghanistan AF AFG 1998 30We would replace the new_sp_m014 with the following four columns:
type sex age n
sp m 014 30This would place each variable in its own column.
22.4 Gather
Here is a nice illustration from RStudio (which they shared under a CC_BY 4.0 license):
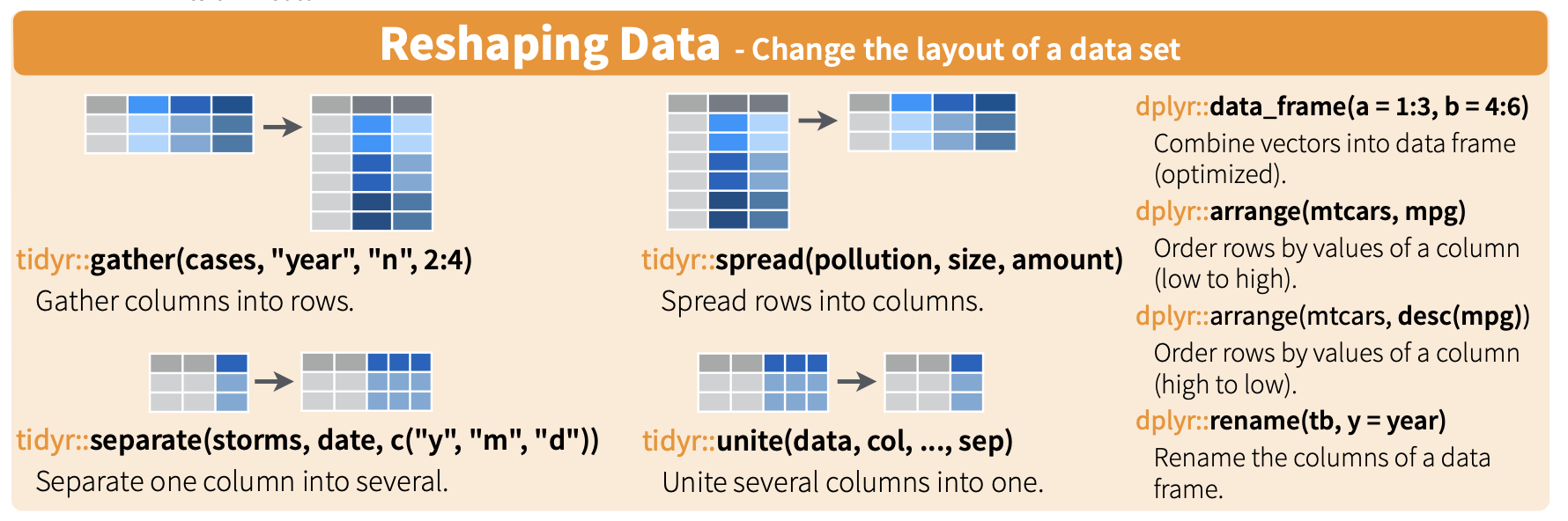
Is the stocks data frame tidy?
stocks <- tibble(
time = as.Date('2009-01-01') + 0:9,
X = rnorm(10, 0, 1),
Y = rnorm(10, 0, 2),
Z = rnorm(10, 0, 4)
)
head(stocks)# A tibble: 6 × 4
time X Y Z
<date> <dbl> <dbl> <dbl>
1 2009-01-01 1.76 -1.34 8.36
2 2009-01-02 1.13 1.49 -1.61
3 2009-01-03 0.445 0.872 -1.14
4 2009-01-04 1.50 -4.74 1.57
5 2009-01-05 -0.602 -2.75 5.75
6 2009-01-06 0.793 0.226 5.42The stocks data frame is not tidy because the variable stock is encoded in the column names.
We can use gather() or pivot_longer() to make it tidy.
stocks %>% gather("stock", "price", -time) %>% head()# A tibble: 6 × 3
time stock price
<date> <chr> <dbl>
1 2009-01-01 X 1.76
2 2009-01-02 X 1.13
3 2009-01-03 X 0.445
4 2009-01-04 X 1.50
5 2009-01-05 X -0.602
6 2009-01-06 X 0.79322.5 Pivot_longer
The gather() function has been superseded and instead it is now recommended to use pivot_longer(), which is easier to use.
Here’s a nice illustration from “Intermediate Reproducible Research in R” (which they shared under a CC_BY 4.0 license), where they caption this figure as follows:
“Pivot longer in tidyr. New columns are called name and value. Notice how the values in A and B columns are stacked on top of each other in the newly created V column.”
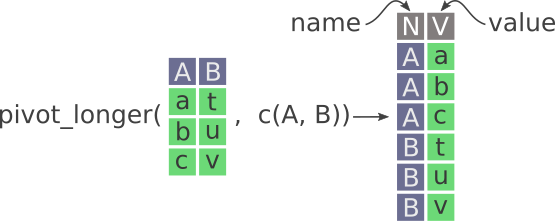
For more detail, see
https://r-cubed-intermediate.rostools.org/sessions/pivots
stocks %>% pivot_longer(c(X,Y,Z), names_to= "stock", values_to = "price") %>%
head()# A tibble: 6 × 3
time stock price
<date> <chr> <dbl>
1 2009-01-01 X 1.76
2 2009-01-01 Y -1.34
3 2009-01-01 Z 8.36
4 2009-01-02 X 1.13
5 2009-01-02 Y 1.49
6 2009-01-02 Z -1.6122.6 WHO TB data
Question: How would we convert this to tidy form?
head(who[,1:6] %>% filter(!is.na(new_sp_m014)))# A tibble: 6 × 6
country iso2 iso3 year new_sp_m014 new_sp_m1524
<chr> <chr> <chr> <dbl> <dbl> <dbl>
1 Afghanistan AF AFG 1997 0 10
2 Afghanistan AF AFG 1998 30 129
3 Afghanistan AF AFG 1999 8 55
4 Afghanistan AF AFG 2000 52 228
5 Afghanistan AF AFG 2001 129 379
6 Afghanistan AF AFG 2002 90 476who.long <- who %>% pivot_longer(starts_with("new"), names_to = "demo", values_to = "n") %>% filter(!is.na(n))
head(who.long)# A tibble: 6 × 6
country iso2 iso3 year demo n
<chr> <chr> <chr> <dbl> <chr> <dbl>
1 Afghanistan AF AFG 1997 new_sp_m014 0
2 Afghanistan AF AFG 1997 new_sp_m1524 10
3 Afghanistan AF AFG 1997 new_sp_m2534 6
4 Afghanistan AF AFG 1997 new_sp_m3544 3
5 Afghanistan AF AFG 1997 new_sp_m4554 5
6 Afghanistan AF AFG 1997 new_sp_m5564 2Question: How would we split demo into variables?
head(who.long)# A tibble: 6 × 6
country iso2 iso3 year demo n
<chr> <chr> <chr> <dbl> <chr> <dbl>
1 Afghanistan AF AFG 1997 new_sp_m014 0
2 Afghanistan AF AFG 1997 new_sp_m1524 10
3 Afghanistan AF AFG 1997 new_sp_m2534 6
4 Afghanistan AF AFG 1997 new_sp_m3544 3
5 Afghanistan AF AFG 1997 new_sp_m4554 5
6 Afghanistan AF AFG 1997 new_sp_m5564 2Look at the variable naming scheme:
names(who) %>% grep("m014",., value=TRUE)[1] "new_sp_m014" "new_sn_m014" "new_ep_m014" "newrel_m014"Question: How should we adjust the demo strings so as to be able to easily split all of them into the desired variables?
who.long <- who.long %>%
mutate(demo = str_replace(demo, "newrel", "new_rel"))
grep("m014",who.long$demo, value=TRUE) %>% unique()[1] "new_sp_m014" "new_sn_m014" "new_ep_m014" "new_rel_m014"Question: After adjusting the demo strings, how would we then separate them into the desired variables?
Hint: Use separate_wider_position() and separate_wider_delim().
who.long.v1 <- who.long %>%
separate(demo, into = c("new", "type", "sexagerange"), sep="_") %>%
separate(sexagerange, into=c("sex","age_range"), sep=1) %>%
select(-new)
head(who.long.v1)# A tibble: 6 × 8
country iso2 iso3 year type sex age_range n
<chr> <chr> <chr> <dbl> <chr> <chr> <chr> <dbl>
1 Afghanistan AF AFG 1997 sp m 014 0
2 Afghanistan AF AFG 1997 sp m 1524 10
3 Afghanistan AF AFG 1997 sp m 2534 6
4 Afghanistan AF AFG 1997 sp m 3544 3
5 Afghanistan AF AFG 1997 sp m 4554 5
6 Afghanistan AF AFG 1997 sp m 5564 2Note that separate() has been superseded in favour of separate_wider_position() and separate_wider_delim(). So here we use those two functions instead of separate():
who.long.v2 <- who.long %>%
separate_wider_delim(demo, names = c("new", "type", "sexagerange"), delim="_") %>%
separate_wider_position(sexagerange, widths=c("sex"=1,"age_range"=4), too_few="align_start" ) %>%
select(-new)
head(who.long.v2)# A tibble: 6 × 8
country iso2 iso3 year type sex age_range n
<chr> <chr> <chr> <dbl> <chr> <chr> <chr> <dbl>
1 Afghanistan AF AFG 1997 sp m 014 0
2 Afghanistan AF AFG 1997 sp m 1524 10
3 Afghanistan AF AFG 1997 sp m 2534 6
4 Afghanistan AF AFG 1997 sp m 3544 3
5 Afghanistan AF AFG 1997 sp m 4554 5
6 Afghanistan AF AFG 1997 sp m 5564 222.7 Conclusion
Now our untidy data are tidy.
head(who.long)# A tibble: 6 × 6
country iso2 iso3 year demo n
<chr> <chr> <chr> <dbl> <chr> <dbl>
1 Afghanistan AF AFG 1997 new_sp_m014 0
2 Afghanistan AF AFG 1997 new_sp_m1524 10
3 Afghanistan AF AFG 1997 new_sp_m2534 6
4 Afghanistan AF AFG 1997 new_sp_m3544 3
5 Afghanistan AF AFG 1997 new_sp_m4554 5
6 Afghanistan AF AFG 1997 new_sp_m5564 222.8 Acknowledgment
This exercise was modeled, in part, on this exercise:
https://people.duke.edu/\~ccc14/cfar-data-workshop-2018/CFAR_R_Workshop_2018_Exercisees.html